Abstract
Background: Interleukin-6 (IL-6) is a multifunctional cytokine involved in various cell functions and diseases. Thus far, several IL-6 inhibitors, such as humanized monoclonal antibody have been used to block excessive IL-6 signaling causing autoimmune and inflammatory diseases. However, anti-IL-6 and anti-IL-6 receptor monoclonal antibodies have some clinical disadvantages, such as a high cost, unfavorable injection route, and tendency to mask infectious diseases. While a small-molecule IL-6 inhibitor would help mitigate these issues, none are currently available.
Objective: The present study evaluated the biological activities of identified compounds on IL-6 stimulus.
Methods: We virtually screened potential IL-6 binders from a compound library using INTerprotein’s Engine for New Drug Design (INTENDD®) followed by the identification of more potent IL-6 binders with artificial intelligence (AI)-guided INTENDD®. The biological activities of the identified compounds were assessed with the IL-6-dependent cell line 7TD1.
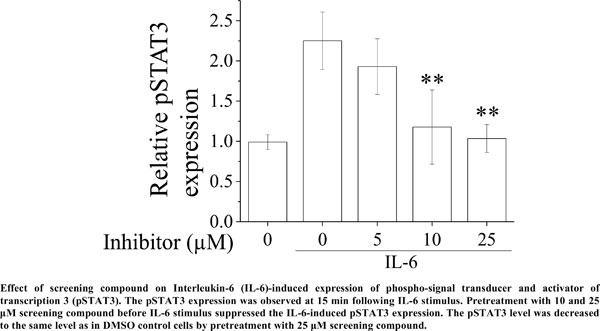
Results: The compounds showed the suppression of IL-6-dependent cell growth in a dose-dependent manner. Furthermore, the identified compound inhibited expression of IL-6-induced phosphorylation of signal transducer and activator of transcription 3 in a dose-dependent manner.
Conclusion: Our screening compound demonstrated an inhibitory effect on IL-6 stimulus. These findings may serve as a basis for the further development of small-molecule IL-6 inhibitors.
Keywords: Artificial intelligence, virtual screening, small molecule inhibitor, interleukin-6, protein-protein interactions, signal transducer and activator of transcription 3.
[http://dx.doi.org/10.1038/324073a0] [PMID: 3491322]
[http://dx.doi.org/10.1084/jem.20081571] [PMID: 19139170]
[http://dx.doi.org/10.1016/j.ajpath.2013.03.014] [PMID: 23673000]
[http://dx.doi.org/10.1101/cshperspect.a016295] [PMID: 25190079]
[http://dx.doi.org/10.7150/ijbs.4614] [PMID: 23136553]
[http://dx.doi.org/10.1155/2014/698313] [PMID: 24524085]
[http://dx.doi.org/10.1016/j.pharmthera.2013.09.004] [PMID: 24076269]
[http://dx.doi.org/10.1038/nrd.2018.45] [PMID: 29725131]
[http://dx.doi.org/10.1093/intimm/dxab011]
[http://dx.doi.org/10.1186/s40164-020-00166-2] [PMID: 32523801]
[http://dx.doi.org/10.2147/TCRM.S3470] [PMID: 19209259]
[http://dx.doi.org/10.1093/rheumatology/ker223] [PMID: 21865281]
[http://dx.doi.org/10.1159/000341698] [PMID: 23108305]
[http://dx.doi.org/10.1042/bj3340297] [PMID: 9716487]
[http://dx.doi.org/10.1016/j.ejca.2005.08.016] [PMID: 16199153]
[http://dx.doi.org/10.1200/JCO.2010.31.8907] [PMID: 22355058]
[http://dx.doi.org/10.2174/092986710792232021] [PMID: 20666726]
[http://dx.doi.org/10.1002/jcp.21567] [PMID: 18767026]
[http://dx.doi.org/10.1016/j.drudis.2018.11.014] [PMID: 30472429]
[http://dx.doi.org/10.1038/s41573-019-0050-3] [PMID: 31801986]
[http://dx.doi.org/10.15252/emmm.202012697] [PMID: 32473600]
[http://dx.doi.org/10.1007/s00401-017-1785-8] [PMID: 29134320]
[http://dx.doi.org/10.1038/s41587-019-0224-x] [PMID: 31477924]
[http://dx.doi.org/10.14533/jbm.19.5]
[http://dx.doi.org/10.1111/j.1365-2567.2007.02679.x] [PMID: 17645497]
[http://dx.doi.org/10.1016/j.leukres.2010.05.011] [PMID: 20542334]
[http://dx.doi.org/10.1093/intimm/dxr077] [PMID: 21937456]
[http://dx.doi.org/10.1002/ibd.20342] [PMID: 18069684]
[http://dx.doi.org/10.1172/JCI64931] [PMID: 23426178]
 114
114 1
1