摘要
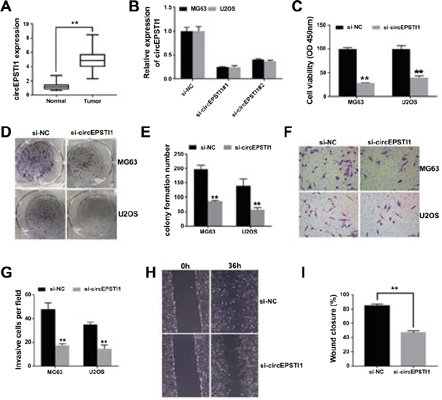
背景:最近的研究报道了环状RNA(circRNA)在肿瘤进展中的重要作用。但是,大多数circRNA在骨肉瘤中的功能和表达情况仍不清楚。 方法:我们通过qRT-PCR检测了circEPSTI1(一种circRNA)在50对配对的正常组织和骨肉瘤组织中的表达。然后,我们进一步探索了circEPSTI1在体外和体内在骨肉瘤进展中的功能。例如,检查了细胞增殖和迁移。进行了一些实验以探索circEPSTI1在miRNA中的调控功能,并研究circEPSTI1在骨肉瘤中的潜在作用。 结果:我们发现circEPSTI1在骨肉瘤中显着上调。 circEPSTI1的抑制抑制了体外骨肉瘤癌细胞的增殖和迁移。双重荧光素酶报告基因检测表明,circEPSTI1和MCL1(髓样细胞白血病1)可以与miR-892b结合,而MCL1和circEPSTI1是miR-892b的靶标。 结论:因此,circEPSTI1-miR-892b-MCL1轴通过miRNA海绵化机制影响骨肉瘤的进展。 circEPSTI1可用作骨肉瘤治疗的靶标和生物标志物。
关键词: circEPSTI1,环状RNA,miR-892b,MCL1,竞争性内源性RNA,骨肉瘤。
[http://dx.doi.org/10.1200/JCO.2014.59.4895] [PMID: 26304877]
[http://dx.doi.org/10.1038/nrc3838] [PMID: 25319867]
[http://dx.doi.org/10.1186/s12943-018-0888-8] [PMID: 30236115]
[http://dx.doi.org/10.1186/s12943-018-0887-9] [PMID: 30236141]
[http://dx.doi.org/10.1016/j.cell.2011.07.014] [PMID: 21802130]
[http://dx.doi.org/10.1158/2159-8290.CD-13-0202] [PMID: 24072616]
[http://dx.doi.org/10.1038/nature12986] [PMID: 24429633]
[http://dx.doi.org/10.1038/nature11928] [PMID: 23446348]
[http://dx.doi.org/10.1038/nature11993] [PMID: 23446346]
[http://dx.doi.org/10.7150/thno.24106] [PMID: 30083277]
[http://dx.doi.org/10.1158/1535-7163.MCT-10-0427] [PMID: 20829195]
[http://dx.doi.org/10.1016/j.pharmthera.2012.09.003] [PMID: 22983152]
[http://dx.doi.org/10.2217/epi-2018-0023] [PMID: 30191736]
[http://dx.doi.org/10.7150/ijbs.27523] [PMID: 30263004]
[http://dx.doi.org/10.7150/ijbs.24360] [PMID: 29559849]
[http://dx.doi.org/10.1016/j.tig.2016.02.001] [PMID: 26922301]
[http://dx.doi.org/10.1016/j.molcel.2018.06.034] [PMID: 30057200]
[http://dx.doi.org/10.1186/s12943-018-0892-z] [PMID: 30285878]
[http://dx.doi.org/10.1016/j.ymthe.2019.01.001]
[http://dx.doi.org/10.1155/2018/7329195] [PMID: 29581984]
[http://dx.doi.org/10.3892/or.2016.5052] [PMID: 27573859]
[http://dx.doi.org/10.1158/0008-5472.CAN-15-1770] [PMID: 26747895]
[http://dx.doi.org/10.1002/jcb.27658] [PMID: 30382592]
[http://dx.doi.org/10.1016/j.tcb.2012.08.011] [PMID: 23026029]
[http://dx.doi.org/10.1038/nature09779] [PMID: 21368834]
[http://dx.doi.org/10.1038/nrd.2016.253] [PMID: 28209992]